Developmental Immunology
Research Focus
General Facts
Research
Selected Publications
Selected Funding, Collaboration
Keywords: Cell Death, Tumor Biology, Non-coding RNAs, Glucocorticoids, Hematopoiesis, Immunology
Research (ÖSTAT Classification) : 301105, 301108, 301902, 301904
Research Focus
Together with their co-workers, IDI investigators explore the basic mechanisms of immune cell development and function, with an emphasis on adaptive immunity, microRNAs and steroid hormones. In addition, they are interested in studying the general principles of cellular transformation, focusing on the role of BCL2-regulated cell death and the p53 signalling network as a barrier to malignant disease. For further details, please see https://www.villungerlab.com.
General Facts
The institute forms part of the Biocenter (https://biocenter.i-med.ac.at) and is organised in four research groups, which are run by Head Professor Andreas Villunger and Associate Professors Jan Wiegers, Verena Labi and Sebastian Herzog. The local resources and infrastructure available include ample modern laboratory space, preclinical models of malignant disease, immune-deficiency and autoimmunity, S2-tissue culture for lenti-or retroviral transduction of model cell lines, histology support, cell sorting (BD FACS_ARIAIII), multicolour flow cytometry (BD FACS-LSR-Fortessa), live-cell imaging (IncuCyte) and a CRISPR/Cas9-based screening platform. The Centre for Molecular Medicine (CeMM) at the Austrian Academy of Science in Vienna, where Andreas Villunger holds an adjunct PI position, also provides swift access to PLACEBO drug-screening and the biomedical sequencing facility (BSF) for RNAseq studies (https://cemm.at).
All faculty members contribute to the teaching and training of students enrolled in human medicine under life sciences at Bachelor’s and Master’s level. A structured PhD programme ensures a first-class training environment for the PhD students in the institute (https://phd-school.i-med.ac.at/). Scientific exchange is guaranteed at various levels, including in regular group meetings, institute and department seminars, as well as participation at international conferences.
Research
The Cell Death Signalling Laboratory
Andreas Villunger
In the Cell Death Signalling laboratory, which is run by the head of the institute, Andreas Villunger, we are currently exploring crosstalk of cell death and cell cycle machinery. As such, our focus is on understanding signalling events that define thresholds for cell death or survival after mitotic errors. Two aspects keep us busy: how mitotic errors are communicated to the BCL2 family network that controls mitochondrial apoptosis; and how p53, a major tumour suppressor, is involved in this context. Within these studies, we have shown that mitotic arrest drives BAX/BAK-mediated cell death through degradation of anti-apoptotic MCL1 and increased abundance of pro-apopotic BIM and NOXA (Fig. 1). In the wake of this finding, we are now investigating how the abundance of these proteins is controlled in mitotic cells.
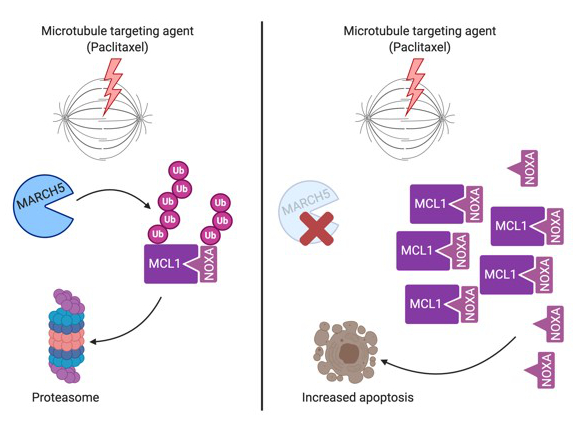
Fig. 1: The mitochondria resident E3 ligase MARCH5 controls turnover of MCL1/NOXA complexes, to regulate cell death in response to spindle poisons.
Moreover, we are investigating the role of a multi-protein signalling complex known as the PIDDosome, which drives p53 activation upon cytokinesis failure (Fig. 2), a process that can prime cells for transformation but that may also represent the desired outcome during normal organ development, as exemplified in the liver or heart.
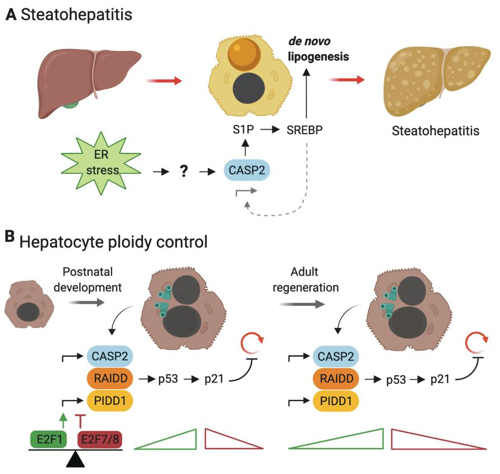
Fig. 2: The PIDDosome and caspase-2 control liver ploidy in early neonatal organogenesis and during E2F-regulated liver regeneration.
The Hormone and Immunity Laboratory
Jan Wiegers
The synthetic derivatives of our stress hormones, glucocorticoids, have successful clinical use as anti-inflammatory and immunosuppressive agents. This suggests that the production of our own (endogenous) glucocorticoids may be inadequate under pathological conditions. Our group focuses on examining the effects of endogenous glucocorticoids on different cells in the immune system, under basal and pathological conditions. Glucocorticoids act primarily through the glucocorticoid receptor (GR), a member of the nuclear receptor family that shows ubiquitous expression. Together with others, we have shown that the critical effects of glucocorticoids are mediated in a manner that is dependent upon cell-type and experimental disease model (Fig. 3). Recently, we reported that the GR is critical to Foxp3+ regulatory T-cell (Treg cell) function in experimental inflammatory colitis, which suggests that endogenous glucocorticoids strengthen Treg cell function in the intestine.
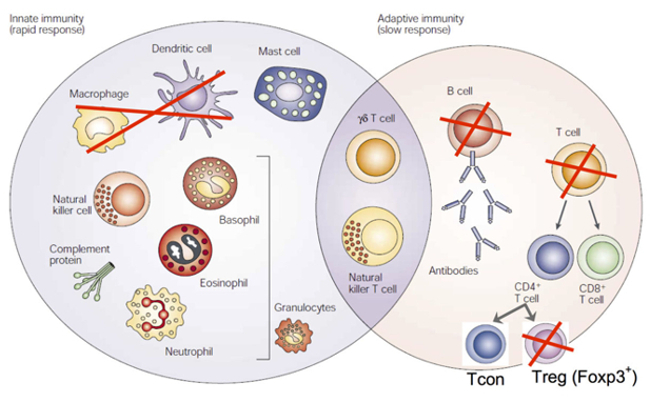
Fig. 3: Immune cell-specific targeting of the GR. Global loss of the GR is lethal. Red crosses show the cell types in which the GR has been specifically deleted to date. Macrophages + dendritic cells: lethal after infection with parasites or injection with superantigen. T-cells (total): lethal upon treatment with polyclonal T-cell activating agents. Foxp3+ Treg cells: defective suppression of experimental inflammatory colitis. B-cells: unknown.
We have also generated a mouse model with selective loss of the GR in B-cells, in order to study the role of glucocorticoids in B-cell development and function (antibody synthesis, immune response regulation). Our aim is to find target cells that are critical for disease model-dependent glucocorticoid therapy. This requires development of a therapeutic strategy, to deliver glucocorticoids in a cell-specific manner. One major advantage of such an approach is a reduction in the strong side-effects of these hormones on other tissues, such as bone (osteoporosis) and muscle (atrophy).
The Experimental Immunology Laboratory
Verena Labi
Pathologies of the immune system represent a major threat to healthy ageing. We study molecular control mechanisms of cell fitness and fate decisions, with a major focus on immune cells and their haematopoietic progenitors. Through genetic cell cultures and mouse models, we aim to understand normal haematopoiesis and immunity in comparison with pathological conditions such as leukaemia, lymphoma, immunodeficiency and autoimmunity. The resulting mechanistic insights should aid the prediction and prevention of disorders and lead to novel strategies for therapeutic intervention. Past achievements include work on:
- Apoptosis as a barrier to autoimmunity and cancer – BCL2 family members BMF and BIM act in synergy to promote apoptosis in the developing embryo and the adult immune system.
- Apoptosis impairs graft quality and transplant outcome – BMF and BIM are rate-limiting for haematopoietic progenitor cell survival, with broader implications for transplantation therapy.
- DNA damage-signalling in B cells – Through fine-tuning of CHK1 dose, antigen-activated B cells exert control over the DNA damage response (Fig. 4).
- Active DNA demethylation shapes B cell identity and function – A combined lack of TET2 and TET3 in B cells prevents enhancer DNA hypomethylation, corrupts gene expression associated with B cell identity and impairs antibody-mediated immunity.
- A microRNA cluster controls B cell fitness and pathology – A PI3K-driven Myc/miR-17~92/Pten autostimulatory loop controls selection of B cell progenitors to the mature B cell pool.
- The binding of miR-17~92 to a single target gene is essential for life – The regulation of BIM transcript levels by miR-17~92 microRNAs serves as a rheostat, which controls cell survival in the developing lung (Fig. 5).
- A paradox, apoptosis fostering tumorigenesis – Classically, the ability of cells to undergo apoptosis is considered a major barrier to cancer. In contrast with popular opinion, we found that apoptosis can promote cancer in the haematopoietic and immune system.
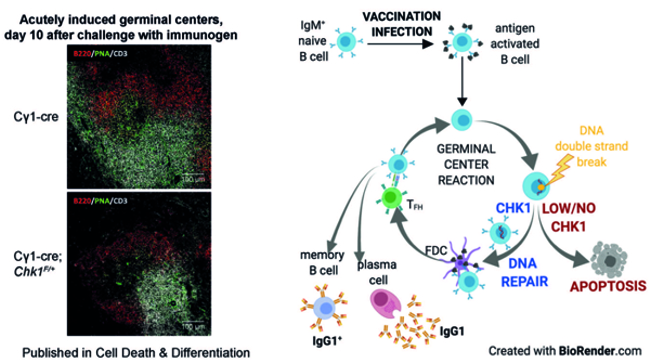
Fig. 4: Immunofluorescence of spleens derived from immunised wild-type mice and animals that lack one allele of DNA damage-response gene CHK1. Germinal centre B cells are green, naïve B cells are red, and T cells are white. The graphical abstract illustrates that effective generation of germinal centres and subsequent antibody-mediated immunity depend on optimal CHK1 dosage.
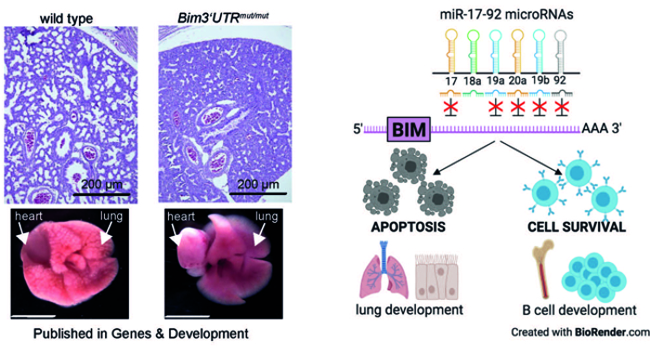
Fig. 5: H&E stains of E19.5 lungs derived from wild-type mice and animals with miR-17-92:BIM interaction deficiency. The latter mice present with hypoplastic lungs when born and are unable to inflate the organ sufficiently. The graphical abstract shows that functional miR-17-92:BIM interactions are dependent on cell context.
The Molecular Gene Regulation Laboratory
Sebastian Herzog
With the work of large consortia, such as FANTOM (functional annotation of the mammalian genome) and ENCODE (encyclopaedia of DNA elements), our understanding of the human genome has dramatically changed. Initially considered as “junk” DNA, it is now clear that the non-protein coding part, which comprises about 98% of the ~3x109 DNA bases, is extensively transcribed and gives rise to numerous non-coding RNAs. Although we are just beginning to understand the complexity of their biological function, growing evidence suggests that at least some non-coding RNAs contribute significantly to the orchestration and fine-tuning of gene expression in both health and disease.
In our group, we are interested in deciphering the role of one type of such non-coding RNAs, microRNAs (miRNAs), in the regulation of complex transcriptional networks, using haematopoiesis and immune function as experimental model systems. MicroRNAs (miRNAs) are small, non-coding RNAs that mediate post-transcriptional silencing of a predicted 60% of protein-coding genes in mammals. Although discovered by Ambros and Ruvkun only in 1993, they have quickly emerged as central mediators of many, if not all, biological processes. In our work, we are trying to decipher how microRNAs influence lymphocyte development and function or, if aberrantly expressed, contribute to diseases such as cancer (Fig. 6). To elucidate the role of individual miRNAs under physiological as well as pathological conditions, we combine gain and loss-of-function approaches, both in in vitro systems and in gene-modified mouse models, with cell biological, biochemical and molecular approaches.
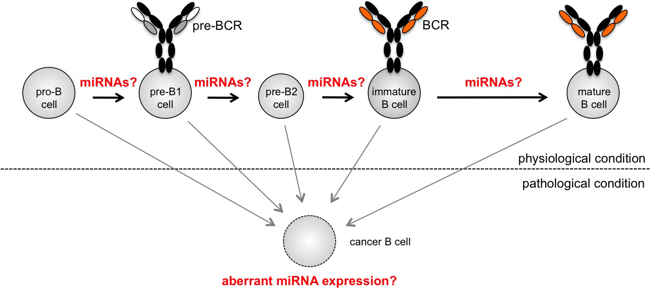
Fig. 6: Early B lymphocyte development is frequently used as a model system for complex transcriptional regulation. Several microRNAs have been linked to defined stage progressions, and others have been shown to promote oncogenic transformation when aberrantly expressed. However, with more than 100 microRNAs expressed in lymphocytes, their individual contribution to cellular function and how they are integrated into transcriptional networks is mostly unclear.
As an additional research interest, we have recently started to focus on the molecular regulation of miRNA biogenesis. Understanding these processes is of key importance, since alterations in the complex processing steps that cut the miRNA from a stem-loop structure on a primary transcript can massively change mature miRNA levels and thus their activity – with severe consequences in several miRNA-mediated malignancies. Experimentally, we are combining genome-wide CRISPR/Cas9-mediated loss-of-function approaches with a broad range of molecular and biochemical approaches, to gain mechanistic insights into how mature miRNAs are generated.
Selected Publications
- Checkpoint kinase 1 is essential for fetal and adult hematopoiesis. Schuler F, Afreen S, Manzl C, Häcker G, Erlacher M, Villunger A., EMBO Rep. 2019. Aug;20(8): e47026.
- MARCH5-dependent degradation of MCL1/NOXA complexes defines susceptibility to antimitotic drug treatment. Haschka MD, Karbon G, Soratroi C, O'Neill KL, Luo X, Villunger A., Cell Death Differ. 2020. 27: 2297–2312.
- E2F-Family Members Engage the PIDDosome to Limit Hepatocyte Ploidy in Liver Development and Regeneration. Sladky VC, Knapp K, Soratroi C, Heppke J, Eichin F, Rocamora-Reverte L, Szabo TG, Bongiovanni L, Westendorp B, Moreno E, van Liere EA, Bakker B, Spierings DCJ, Wardenaar R, Pereyra D, Starlinger P, Schultze S, Trauner M, Stojakovic T, Scharnagl H, Fava LL, Foijer F, de Bruin A, Villunger A., Dev Cell. 2020. Feb 10; 52(3): 335-349.e7.
- Glucocorticoid Receptor-Deficient Foxp3+ Regulatory T Cells Fail to Control Experimental Inflammatory Bowel Disease. Rocamora-Reverte L, Tuzlak S, von Raffay L, Tisch M, Fiegl H, Drach M, Reichardt HM, Villunger A, Tischner D, Wiegers GJ., Front Immunol. Mar 18; 10: 472.
- Context-specific regulation of cell survival by a miRNA-controlled BIM rheostat. Labi V, Peng S, Klironomos F, Munschauer M, Kastelic N, Chakraborty T, Schoeler K, Derudder E, Martella M, Mastrobuoni G, Hernandez-Miranda LR, Lahmann I, Kocks C, Birchmeier C, Kempa K, Quintanilla-Martinez de Fend L, Landthaler M, Rajewsky N, Rajewsky K., Genes Dev. 2019. Dec 1; 33(23-24): 1673-87.
- CHK1 dosage in germinal center B cells controls humoral immunity. Schoeler K, Jakic B, Heppke J, Soratroi C, Aufschnaiter A, Hermann-Kleiter N, Villunger A, Labi V., Cell Death Differ. 2019. Dec; 26(12): 2551-67.
- Differential roles of miR-15a/16-1 and miR-497/195 clusters in immune cell development and homeostasis. Hutter K, Rülicke T, Drach M, Andersen L, Villunger A, Herzog S., FEBS J. 2020 Jul 23.
- SAFB2 Enables the Processing of Suboptimal Stem-Loop Structures in Clustered Primary miRNA Transcripts. Hutter K, Lohmüller M, Jukic A, Eichin F, Avci S, Labi V, Szabo TG, Hoser SM, Hüttenhofer A, Villunger A, Herzog S., Mol Cell. 2020 Jun 4; 78(5): 876-889.e6.
Selection of Funding
- POLICE - The PIDDosome in Centrosome and Ploidy-Surveillance, ERC, Andreas Villunger
- Cell death control on extended mitotic arrest, FWF, Andreas Villunger
- miRNA:mRNA interactions at the level of a single target gene, FWF, Verena Labi
- Unraveling miR-15 function in health and disease, FWF, Sebastian Herzog
Collaborations
- Alain de Bruin, Utrecht University, The Netherlands
- Floris Foijer, ERIBA, Groningen, The Netherlands
- Ana Garcia-Saez, University of Cologne, Germany
- Luca Fava, CIBIO, Trento, Italy
- Klaus Rajewsky, Max-Delbrück Center for Molecular Medicine, Berlin, Germany
- Mir-Farzin Mashreghi, DRFZ, Berlin, Germany
- Miriam Erlacher, Univ. Hospital, Freiburg, Germany
Devices & Services
BD LSRFortessa Cell Analyzer; BD FACS ARIA III Cell Sorter; Attune NxT Cell Analyzer; IncuCyte S3 Live-Cell Analysis System; CRISPR/Cas9-based screening service
 Univ.-Prof. Mag. Dr.rer.nat. Andreas Villunger
Univ.-Prof. Mag. Dr.rer.nat. Andreas Villunger
Director
Contact:
Innrain 80-82
6020 Innsbruck
Austria
Email: andreas.villunger@i-med.ac.at
Phone: +43 512 9003 70830
Fax: +43 512 9003 73960
www.villungerlab.com



