Genetic Epidemiology
Research Focus
General Facts
Research
Selected Publications
Selected Funding, Collaboration
Devices & Services
Keywords: Genetic epidemiology, lipoprotein metabolism, complex phenotypes, genome-wide association studies, mitochondrial DNA, computational genetics, genotype imputation, risk factors, kidney disease, cardiovascular disease
Research (ÖSTAT Classification) : 102004,102010, 301301, 301302, 303007
Research Focus
We aim to identify determinants of health and disease in relation to genetic variability, environmental components and biochemical parameters and to study their physiological or pathophysiological functions. Our phenotypes of interest are complex in nature, owing to their interactions, and they relate to atherosclerosis, diabetes mellitus, metabolic syndrome, kidney disease, cancer and associated intermediate phenotypes such as lipoprotein metabolism and inflammation.
General Facts
The institute, which has about 18 – 20 members (Fig. 1), serves as a bridge between basic and clinical research. We have three pillars, the strength of which lies in interconnected collaboration: 1) a protein chemistry and cell culture laboratory undertakes a variety of structural-functional and epidemiological studies into various phenotypes related to lipoprotein metabolism and other metabolic phenotypes (e.g. Lp(a), apolipoprotein A-IV, afamin, cholesterol efflux assays); 2) a molecular genetics laboratory performs sequencing and genotyping for various projects, with a strong focus on mitochondrial DNA as well as complex genetic regions such as the K-IV type 2 region of the LPA gene – new technologies such as nanopore sequencing have recently became a major focus of this research; 3) the computational and statistical genetics laboratory focusses on statistics, epidemiology, computer science and bioinformatics and represents an important link between the various research groups. A close interconnection of these three “units” to form a strongly collaborative alliance has become increasingly evident in recent years. There are barely any major projects that involve fewer than two of these main groups. The output and success of the institute are based upon constant dialogue between the various disciplines, in a problem-oriented and critical elucidation of research questions.

Fig. 1: Genetic Epidemiology team. The picture shows all members of staff in 2019/20.
Research
Lipoprotein(a)
Stefan Coassin, Sebastian Schönherr, Claudia Lamina, Florian Kronenberg
High concentrations of lipoprotein(a) [Lp(a)] are a strong risk factor for cardiovascular disease. These high concentrations are mostly genetically influenced by the LPA gene. Despite this strong genetic regulation, Lp(a) concentrations in individuals with the same isoform combination can vary 200-fold. This suggests that Lp(a) levels are modified by additional genetic variants, which have to be identified. However, the LPA gene contains a large, hypervariable region, which is not properly covered by any genome reference database. It can, however, encompass up to 70% of the gene and may therefore contain most of the genetic variability in LPA. We pursue two main research aims.
- Identification of new genetic variants regulating Lp(a)
A major part of our work concerns the investigation of mutations in the KIV-2 repeat region of the LPA gene. By combining the computational expertise of the institute in the detection of low-level mutations and our long-standing experience in Lp(a) research, we have developed a sequencing approach that is capable of resolving the variability in this large region. A screen of 123 individuals revealed an unprecedented level of genetic variability, with several hundred novel variants, some of which strongly regulate Lp(a) concentrations (J Lipid Res 2019; Genome Medicine 2020) (Fig. 2). Several variants identified in the KIV-2 region as well as the rest of the LPA gene are now characterised by genetic and functional analyses to improve understanding of the genetic architecture of Lp(a).
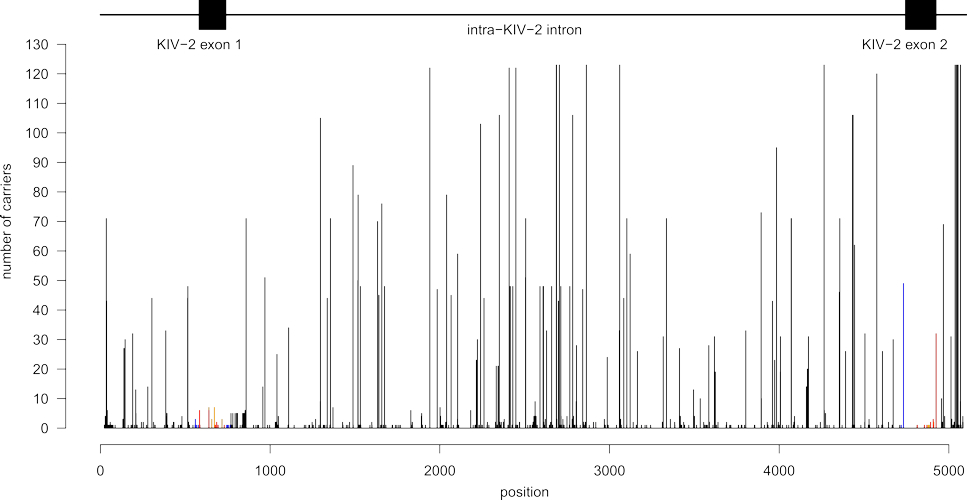
Fig. 2: Variants hidden in the complex LPA KIV-2 copy number variable region. Variants found when sequencing the KIV-2 region of the LPA gene, the complex “dark region” of the genome, in 123 individuals. On the graph, all mutations found are mapped on a single repeat. Each bar therefore indicates that a mutation was found at the corresponding site in any of the up to 80 repeats that can be present in every individual; the y axis indicates the number of individuals in which the respective mutation was found. Some positions were indeed variable in every single individual sequenced. The exon structure of the KIV-2 repeat is shown above the graph.
- Evaluation of the clinical impact of Lp(a)
Although Lp(a) is well established as a causal risk factor for cardiovascular disease, many questions still deserve further research. Specific Lp(a)-lowering drugs are not yet available but there are promising candidates, which are currently being evaluated in clinical trials. For the planning of such trials, it is of utmost importance to estimate the required Lp(a)-lowering in order to improve clinical outcomes effectively. Using data from five studies including almost 14,000 participants, we estimated that Lp(a) would have to be reduced by 66 mg/dL in order to achieve the same effect on cardiovascular risk reduction as reducing LDL-cholesterol by 1 mmol/L by means of statins (JAMA Cardiol 2019). This would correspond to a risk reduction of 45% over a lifetime or 22% in the short term. This estimation was made possible by applying a method called Mendelian randomisation. With this special statistical technique, it is possible to derive the causal relationship between a risk factor, such as Lp(a), and a disease for which genetic variants that regulate Lp(a) are known.
Apolipoprotein A-IV and Afamin
Barbara Kollerits, Claudia Lamina, Hans Dieplinger, Florian Kronenberg
We are investigating these two proteins in large epidemiological studies that include up to 30,000 individuals. The antiatherogenic apolipoprotein A-IV is strongly related to kidney function and linked to overall mortality, cardiovascular disease and cancer. In the case of afamin, we showed a pronounced link with metabolic syndrome and diabetes mellitus and we are currently investigating its connection with kidney function, cardiovascular disease, iron metabolism and anaemia.
Mitochondrial DNA
Hansi Weissensteiner, Federica Fazzini, Lukas Forer, Monika Summerer, Florian Kronenberg, Sebastian Schönherr
The group has a strong interest in the mitochondrial genome and related research. Specifically, three main topics are covered here.
- Bioinformatics tools
The group successfully develops software to analyse mitochondrial DNA (mtDNA). Based on this genome, which is inherited strictly maternally, individuals can be grouped according their female ancestors. We therefore developed the HaploGrep software, which performs this classification and enjoys strong acceptance by the community. Additionally, we developed mtDNA-Server (https://mtdna-server.uibk.ac.at) in order to analyse next-generation sequencing (NGS) of mtDNA data. Our latest software, Haplocheck, allows fast contamination detection in large NGS sequencing projects, by inspecting polymorphic sites in the mtDNA (Genome Research, 2021). At present, we are working on the integration of our mtDNA software into a central web service (https://mitoverse.i-med.ac.at) as well as constantly updating our software to meet the needs of thousands of researchers worldwide.
- mtDNA cancer research
Mitochondria play a vital role in energy production, and the involvement of mtDNA in cancer formation is therefore of special interest. In cooperation with researchers from Experimental Urology and Cranio-Maxillofacial and Oral Surgery, we analysed low-level mutations in the mtDNA in cancer/benign tissue pairs in the case of prostate cancer (Nature Commun. 2020) and oral squamous cell carcinoma. For both cancer types, we were able to detect accumulations of somatic mutations in the cancer tissues with the aid of NGS. In addition, we described mutational and tissue-specific differences in tumour and benign prostate samples using high-resolution respirometry and RNA sequencing (Fig. 3).
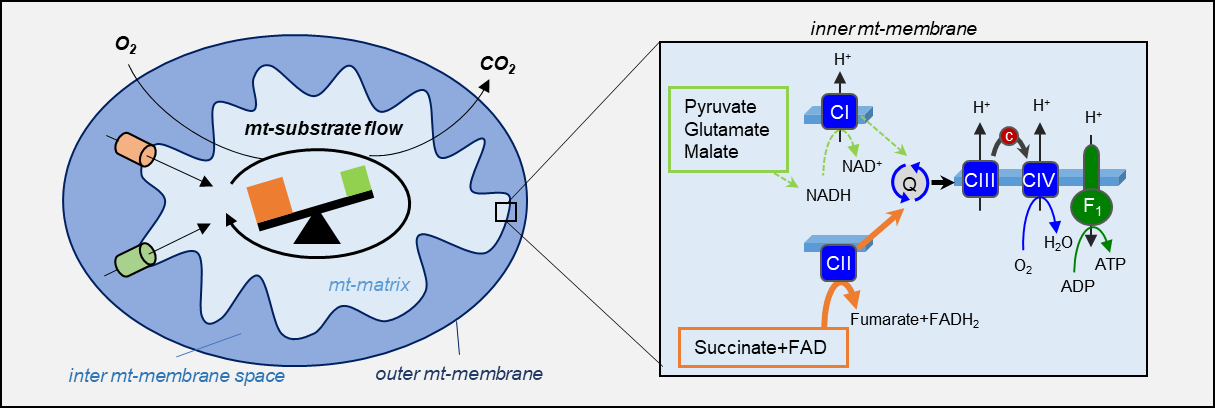
Fig. 3: OXPHOS substrate shift from glutamate and malate to succinate in high-grade prostate cancer tissue.
- Mitochondrial copy number
Mitochondrial content is crucial to the energy demand of a cell type. Changes in the mitochondrial copy number (mtCN) could be disease-related. By refining the method of measuring the mitochondrial copy number using a duplex quantitative PCR-based assay, the group subsequently investigated the mtCN in 4,812 patients from the German Chronic Kidney Disease (GCKD) cohort (Kidney Int. 2019). Additionally, the mtCN was analysed and compared in 2 different cohorts, amounting to a total of over 14,000 samples, as part of the collaboration with the EURAC Bozen and the GCKD-Consortium (J Intern Med. 2021).
Michigan Imputation Server as a Backbone for GWA Studies Worldwide
Sebastian Schönherr, Florian Kronenberg, Lukas Forer
The Michigan Imputation Server (https://imputationserver.sph.umich.edu) (Fig. 4) is an ongoing collaboration with the University of Michigan (USA) and the EURAC research centre (Italy). It provides a free genotype imputation service for genome-wide association studies (GWAS). With >65.2 million imputed genomes, >6,800 active users and >1000 citations of the Nature Genetics paper published in 2016, it is the central backbone for GWAS worldwide. One aim of the Michigan Imputation Server is to provide population-specific reference panels, in order to improve imputation for non-European individuals. For example, the Michigan Imputation Server supports the Genome Asia reference panel, which includes a whole-genome sequencing reference dataset from 1,739 individuals in 219 population groups and 64 countries across Asia and facilitates genetic studies of Asian populations (Nature 2019). Another example is the TOPMed-based imputation reference panel, which includes 97,256 individuals with 308,107,085 SNPs and indels (TOPMed Imputation Server). For COVID-19 related studies, we are also providing a HLA reference panel. All the services and reference panels mentioned are free of charge for academic and non-academic users worldwide.
The current focus of this research project is on the development of new methods for automation of the overall imputation process (e.g. by using an API), on the calculation of genetic risk scores (e.g. via the PGS catalogue) included within each imputation, and on the improvement and maintenance of the Michigan Imputation Server.
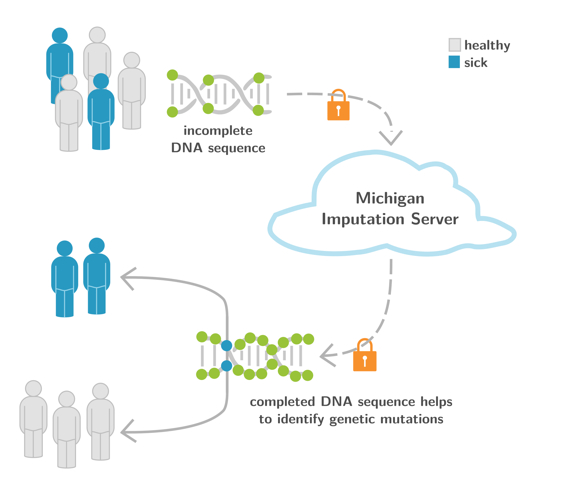
Fig. 4: Michigan Imputation Server Workflow. In a first step, DNA positions derived from microarray chips are uploaded to the server. The uploaded positions are then compared with the complete reference DNA (up to 32,000 samples) and the unknown DNA sections are filled in by means of genotype imputation, in order to produce a DNA sequence that is as complete as possible.
Askimed – The Next-Generation eCRF System for Medical Studies in the Cloud
Lukas Forer, Hansi Weissensteiner, Florian Kronenberg, Sebastian Schönherr
High-quality collected data are key to medical studies or registries. Furthermore, new study formalities require each data point to be collected in a reproducible way and data changes to be tracked within a digital system. In order to support studies with implementing these new demands and to optimise data collection, we have developed a software solution to simplify user study workflows. Askimed (https://www.askimed.com) described in Fig. 5 is a cloud-based web platform that provides an all-in-one solution for medical studies of any size. The main features of Askimed are: (1) data collection based on electronic case report forms (eCRF), (2) data management including a permission system for study collaborators and (3) data preparation for data analysis. Askimed integrates user-friendly tools for data collection, error elimination (e.g. plausibilities) and data security (e.g. security log, audit trail). Using Askimed, researchers can implement clinical studies or registries and focus on the collaborative aspect within a study. Askimed offers a hosted version for our cooperation partners and we currently have over 15 ongoing collaborations within the Medical University of Innsbruck (e.g. Endometriosis Registry, ACEI-COVID Study, FH Registry). In recent years, we have also been involved in many international consortia and large studies. We have also digitally accumulated over 150,000 interviews and helped our partners to optimise their data collection and workflows.
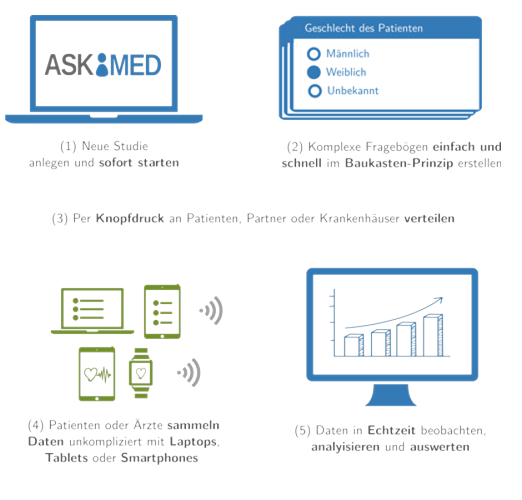
Fig. 5: The Askimed platform (https://www.askimed.com) is a next generation eCRF system for medical studies in the cloud. It includes all the features required to conduct state-of-the-art clinical trials and provides tools for CRF creation, data collection and data analysis of large datasets.
Mutation Analysis of Complex Genome Regions and Nanopore Sequencing
Stefan Coassin, Hansi Weissensteiner, Sebastian Schönherr
We investigate genome regions with complex structures by developing new computational approaches and by exploring nanopore sequencing applications. We use the findings from the projects that cover the KIV-2 region or the mutation analysis of mtDNA to devise more general approaches to tackling complex genome regions in available short-read sequencing data, such as the publicly available UK Biobank. This resource contains genetic and phenotypical data from 500,000 individuals across the United Kingdom. At the moment, we leverage this data to search the available 200,000 exome sequencing datasets for new genetic variants in the complex KIV-2 region described above and to search for novel loci associated with Lp(a) levels using GWAS approaches.
Long-read nanopore sequencing allows sequencing through repetitive or complex genome structures, direct phasing of mutations located far apart, direct determination of chemically modified bases, and even targeted sequencing by selection of relevant reads from the sequencing data stream in real-time. Using this technology, we were recently able contribute to the resolution of a very complex gene in the previously unsequenced organism Minona ileanae (in collaboration with Peter Ladurner, Institute of Zoology, University of Innsbruck; Pjeta et al, Philos Trans R Soc Lond B Biol Sci, 2019) and to elucidate the structure of a large duplication in transgenic mice (in collaboration with Katrin Watschinger and Ernst Werner, Institute of Biological Chemistry).
Structural Biology of Therapeutic Protein Targets and Drug Development
Bernhard Rupp, Andreas Naschberger
We support multiple interdisciplinary projects with detailed 3-dimensional molecular structures of disease-related proteins, by means of experimental methods of X-ray diffraction, cryo-EM and in-silico modelling. FWF-funded X-ray studies of the plasma glycoprotein afamin have provided evidence of lipid and MRI contrast transport, and TLR3 receptor-biglycan complex modelling has revealed potential therapeutic pathways to prevent cardiovascular calcification. Structural analysis of the mutations in mitochondrial complex I has revealed implications for prostate tumour progression and treatment. Analysis, validation and re-refinement of SARS-Cov2 target protein structure complexes with drug lead compounds are helping the international research community with the development of novel antiviral therapies.
HOROS: Host Response in Opportunistic Infections
Stefan Coassin, Claudia Lamina, Florian Kronenberg
The group is a part of this FWF-funded doctoral programme and we focus on high-risk populations, such as patients with chronic kidney disease (CKD), that are highly susceptible to recurrent opportunistic infections. In the German Chronic Kidney Disease (GCKD) study, a prospectively followed cohort of 5217 patients with impaired kidney function, we recently described strong connections of mitochondrial DNA copy number (Kidney Int. 2019) and relative telomere length (Kidney Int. 2020) with mortality, cardiovascular disease and infections. In ongoing projects, we are investigating cholesterol efflux as well as PCSK9 concentrations in the same cohort, and in patients with peripheral arterial disease, we have shown that Lp(a) and PCSK9 independently contribute to the risk of peripheral arterial disease (Atherosclerosis 2021).
Selected Publications
- Di Maio S, Grüneis R, Streiter G, Lamina C, Maglione M, Schoenherr S, Öfner D, Thorand B, Peters A, Eckardt K-U, Köttgen A, Kronenberg F, Coassin S: Investigation of a nonsense mutation located in the complex KIV-2 copy number variation region of apolipoprotein(a) in 10,910 individuals. GENOME MEDICINE: 2020; 12: 74
- Coassin S*, Schönherr S*, Weissensteiner H, Erhart G, Forer L, Losso JL, Lamina C, Haun M, Utermann G, Paulweber B, Specht G, Kronenberg F: A comprehensive map of single base polymorphisms in the hypervariable LPA Kringle IV-2 copy number variation region. JOURNAL OF LIPID RESEARCH: 2019; 60:186-199; * authors contributed equally
- Lamina C, Kronenberg F: Estimation of the Required Lipoprotein(a)-Lowering Therapeutic Effect Size for Reduction in Coronary Heart Disease Outcomes: A Mendelian Randomization Analysis. JAMA CARDIOLOGY: 2019; 4:575-579
- Schöpf B, Weissensteiner H, Schäfer G, Fazzini F, Charoentong P, Naschberger A, Rupp B, Fendt L, Bukur V, Giese I, Sorn P, Sant'Anna-Silva AC, Iglesias-Gonzalez J, Sahin U, Kronenberg F, Gnaiger E, Klocker H: OXPHOS remodeling in high-grade prostate cancer involves mtDNA mutations and increased succinate oxidation. NATURE COMMUNICATIONS: 2020; 11:1487
- Fazzini F, Lamina C, Fendt L, Schultheiss UT, Kotsis F, Hicks AA, Meiselbach H, Weissensteiner H, Forer L, Krane V, Eckardt KU, Köttgen A, Kronenberg F: Mitochondrial DNA copy number is associated with mortality and infections in a large cohort of patients with chronic kidney disease. KIDNEY INTERNATIONAL 2019; 96:480-488
- GenomeAsia100K Consortium: The GenomeAsia 100K Project enables genetic discoveries across Asia. NATURE: 2019; 576:106-111
- Naschberger A, Juyoux P, von VJ, Rupp B, Bowler MW: Controlled dehydration, structural flexibility and gadolinium MRI contrast compound binding in the human plasma glycoprotein afamin. ACTA CRYSTALLOGRAPHICA SECTION D BIOLOGICAL CRYSTALLOGRAPHY: 2019; 75:1071-1083
- Pjeta R, Wunderer J, Bertemes P, Hofer T, Salvenmoser W, Lengerer B, Coassin S, Erhart G, Beisel C, Sobral D, Kremser L, Lindner H, Curini-Galletti M, Stelzer CP, Hess MW, Ladurner P: Temporary adhesion of the proseriate flatworm Minona ileanae. PHILOSOPHICAL TRANSACTIONS OF THE ROYAL SOCIETY B BIOLOGICAL SCIENCES: 2019; 374:20190194
- Fazzini F, Lamina C, Raschenberger J, Schultheiss UT, Kotsis F, Schönherr S, Weissensteiner H, Forer L, Steinbrenner I, Meiselbach H, Bärthlein B, Wanner C, Eckardt K-U, Köttgen A, Kronenberg F: Results from the German Chronic Kidney Disease (GCKD) study support association of relative telomere length with mortality in a large cohort of patients with moderate chronic kidney disease. KIDNEY INTERNATIONAL: 2020; 98: 488-497
- Koller A, Fazzini F, Lamina C, Rantner B, Kollerits B, Stadler M, Klein-Weigel P, Fraedrich G, Kronenberg F: Mitochondrial DNA copy number is associated with all-cause mortality and cardiovascular events in patients with peripheral arterial disease. JOURNAL OF INTERNAL MEDICINE: 2020; 287:569-579
- Fendt L*, Fazzini F*, Weissensteiner H, Bruckmoser E, Schonherr S, Schafer G, Losso JL, Streiter GA, Lamina C, Rasse M, Klocker H, Kofler B, Kloss-Brandstatter A, Huck CW, Kronenberg F, Laimer J: Profiling of Mitochondrial DNA Heteroplasmy in a Prospective Oral Squamous Cell Carcinoma Study. Cancers (BASEL): 2020; 12:1933; * authors contributed equally
- Coassin S, Kronenberg F: Mechanistic insights into lipoprotein(a): from infamous to 'inflammous'. EUROPEAN HEART JOURNAL: 2020; 41: 2272-2274
- Fazzini F, Lamina C, Raftopoulou A, Koller A, Fuchsberger C, Pattaro C, Del Greco F, Döttelmayer P, Fendt L, Fritz J, Meiselbach H, Schönherr S, Forer L, Weissensteiner H, Pramstaller P, Eckardt, K-U, Hicks A, Kronenberg F: Association of mitochondrial DNA copy number with metabolic syndrome and type 2 diabetes in 14,176 individuals. Journal of Internal Medicine (in press)
- Kheirkhah A, Lamina C, Rantner B, Kollerits B, Stadler M, Pohlhammer J, Klein-Weigel P, Fraedrich G, Kronenberg F: Elevated levels of serum PCSK9 in male patients with symptomatic peripheral artery disease: The CAVASIC study. Atherosclerosis: 2021; 316: 41-47
- Weissensteiner H, Forer L, Fendt L, Kheirkhaha A, Salas A, Kronenberg F, Schoenherr S: Contamination detection in sequencing studies using the mitochondrial phylogeny. Genome Research: 2021 (in press)
Selection of Funding
- "Molecular investigation of high Lp(a) pedigrees", Lp(a)CARE; Stefan Coassin and Florian Kronenberg
- "Apolipoprotein(a) Isoforms and apolipoprotein A-IV concentrations in the Rotterdam Study and DiaGene Study", Erasmus Medical Center; Florian Kronenberg
- "Studies of Rare Genetic Variation in the Isolated Population of Sardinia”, NIH Subaward agreement with the University of Michigan (Goncalo Abecasis); Lukas Forer (together with Sebastian Schönherr, Christian Fuchsberger)
- “Askimed-Cloud-Plattform zur Erhebung, Verwaltung und Analyse von Daten im Gesundheitssektor”, Lukas Forer, Sebastian Schönherr
- "Host response in opportunistic infections", FWF PhD Programme, Florian Kronenberg
- “Studying an Lp(a) mutation in human RNA and liver organoids”, FWF, Stefan Coassin
Collaborations
- Gonçalo Abecasis, Center of Statistical Genetics, University of Michigan, Ann Arbor, USA
- Steven C. Hunt, Cardiovascular Genetics Division, University of Utah, Salt Lake City, USA
- Institute of Epidemiology, Helmholtz Zentrum München, Neuherberg, Germany
- Kai-Uwe Eckardt, Department of Nephrology and Medical Intensive Care, Charité—Universitätsmedizin Berlin, Berlin, Germany
- Nicole Probst-Hensch, Swiss Tropical and Public Health Institute, Basel, Switzerland
- Anna Köttgen, Institute of Genetic Epidemiology, University of Freiburg, Freiburg, Germany
- Eric Sijbrands, Department of Internal Medicine, Erasmus MC-University Medical Center Rotterdam, The Netherlands
- Iris M. Heid, Department of Genetic Epidemiology, University of Regensburg, Regensburg, Germany
- Gilles Lambert, Laboratoire INSERM UMR 1188 DéTROI, Université de La Réunion, Sainte Clotilde, France.
Devices & Services
- Free Next-Generation Genotype Imputation Service using most comprehensive reference genomes: https://imputationserver.sph.umich.edu
- mtDNA-Server & new mtDNA analysis tool sets: https://mitoverse.i-med.ac.at
- Haplogroup classification service HaploGrep2 : https://haplogrep.i-med.ac.at
- Next generation eCRF system for clinical studies: https://www.askimed.com/
 Univ.-Prof. Dr. med. univ. Florian Kronenberg
Univ.-Prof. Dr. med. univ. Florian Kronenberg
Director
Contact:
Schöpfstr. 41
6020 Innsbruck
Austria
Email: FIorian.Kronenberg@i-med.ac.at
Phone: +43 512 9003 70560
Fax: +43 512 9003 73561
http://www3.i-med.ac.at/genepi



